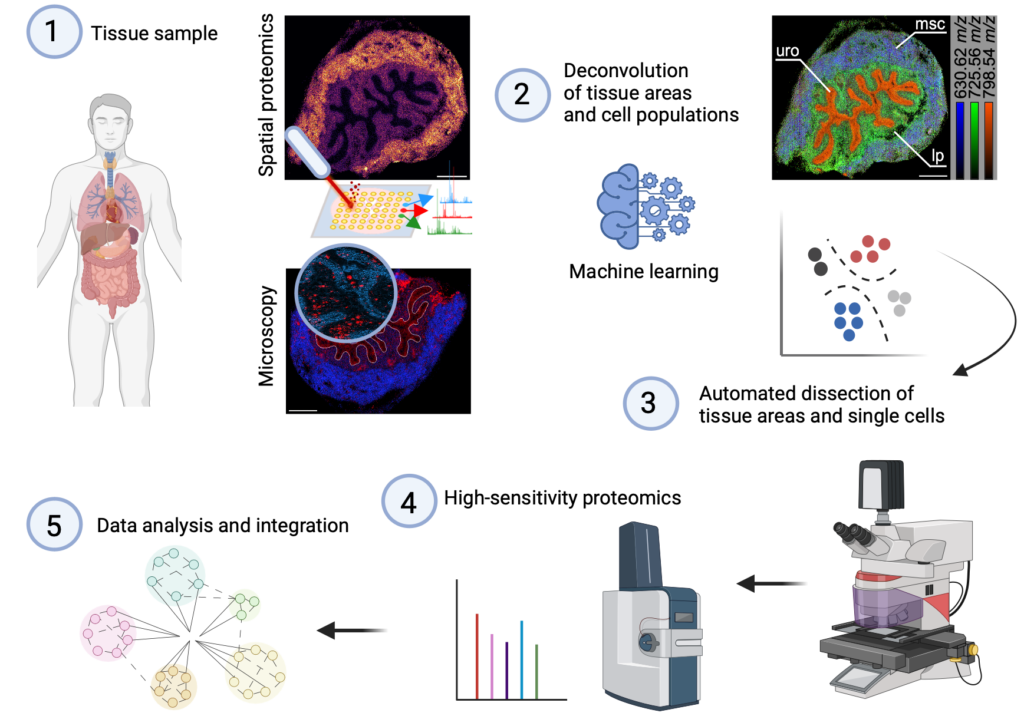
The MOSAIC facility is our interdisciplinary platform for multimodal analysis. It integrates advanced microscopy, mass spectrometry, laser capture microdissection, and high-sensitivity proteomics in a computational framework. By combining state-of-the-art instrumentation with standardized workflows and automated data processing pipelines, MOSAIC enables reproducible, high-resolution mapping of cellular phenotypes and molecular signatures in complex tissue microenvironments and liquid biopsies. The facility provides end-to-end support—from experimental design and sample preparation to data integration and bioinformatic analysis—thereby accelerating translational research in infection, inflammation, and cancer.
MOSAIC aims to unravel the spatial mechanism regulating the crosstalk of immune cells with the surrounding tissue. We use and develope registration algorithms to combine multimodal imaging approaches, such as microscopy and mass spectrometry imaging (1). Artificial intelligence is used to deconvolute the data into tissue- and cell specific signals (2) for automated dissection by laser capture microdissection (3) and high-sensitivity proteomics (4). Data analysis and integration are performed with python-based computing and automation via the snakemake workflow management system (5).
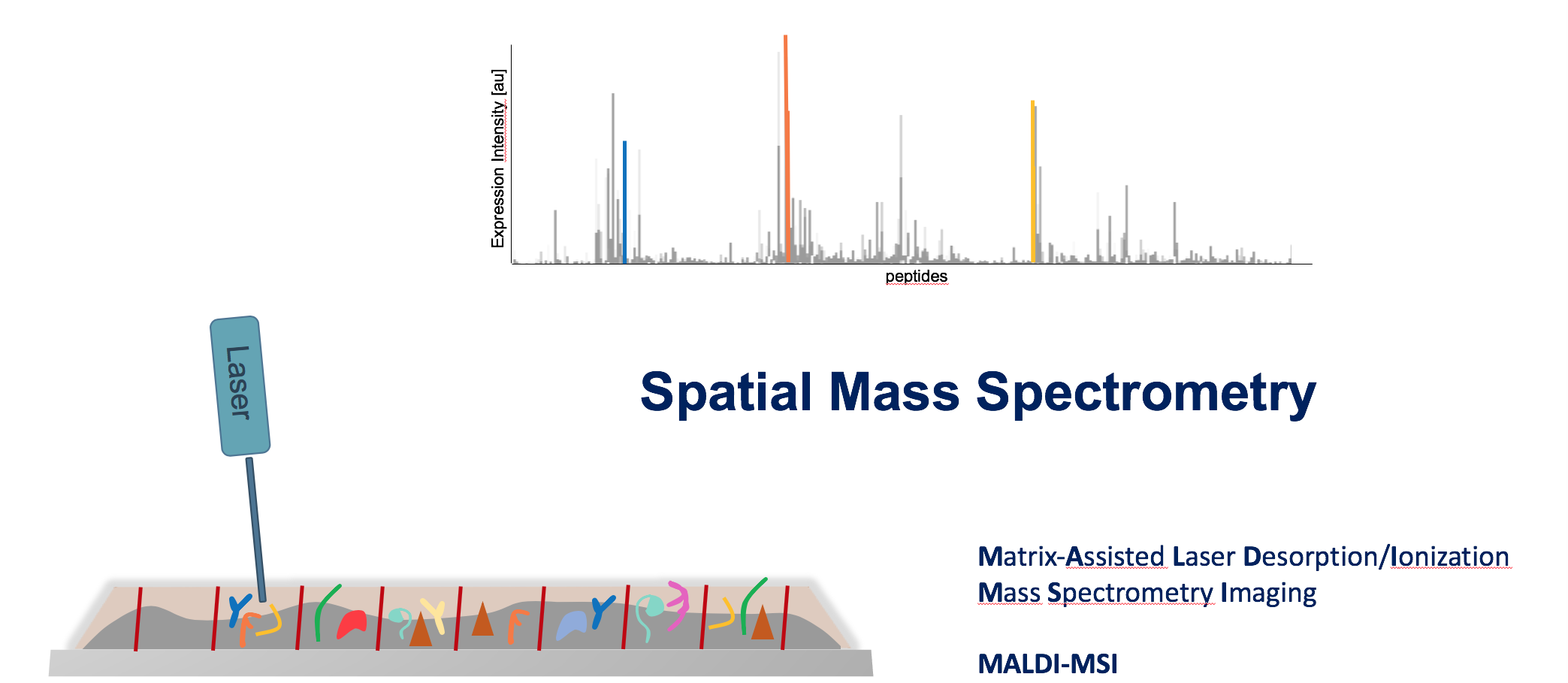
Mass Spectrometry Imaging
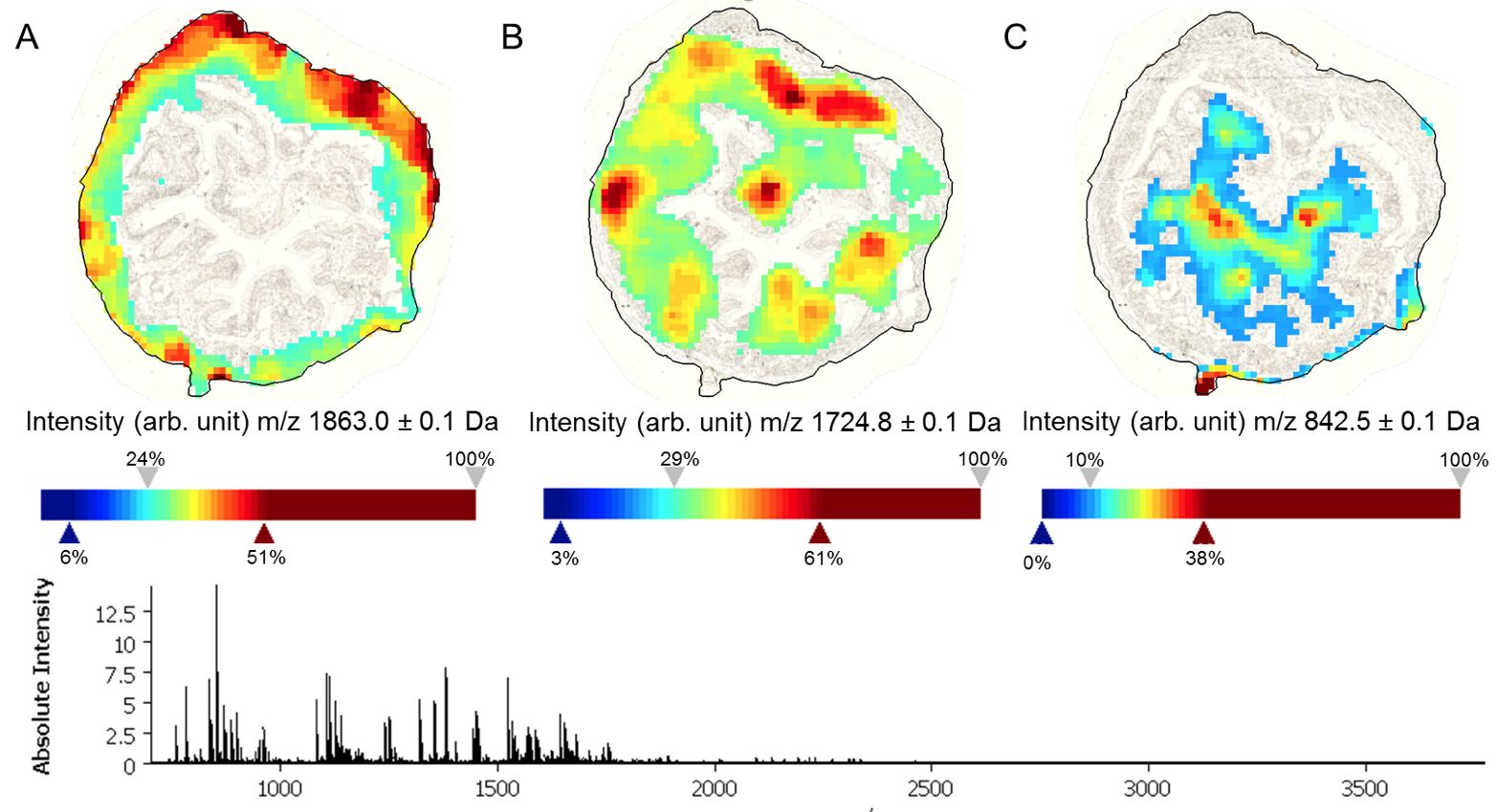
Mass spectrometry imaging (MSI) is a label-free analytical technique that enables the spatially resolved detection of molecules directly within tissue sections. By combining molecular specificity with histological context, MSI allows the visualization of proteins, lipids, metabolites, and drugs without prior knowledge of their distribution. In biomedical research, this approach is particularly powerful for mapping disease-associated molecular changes, characterizing tumor heterogeneity, studying immune–tissue interactions, and tracking therapeutic compounds within organs. As a result, MSI bridges molecular biology and spatial pathology, providing insights into tissue organization and functional microenvironments that cannot be obtained from bulk analyses alone.
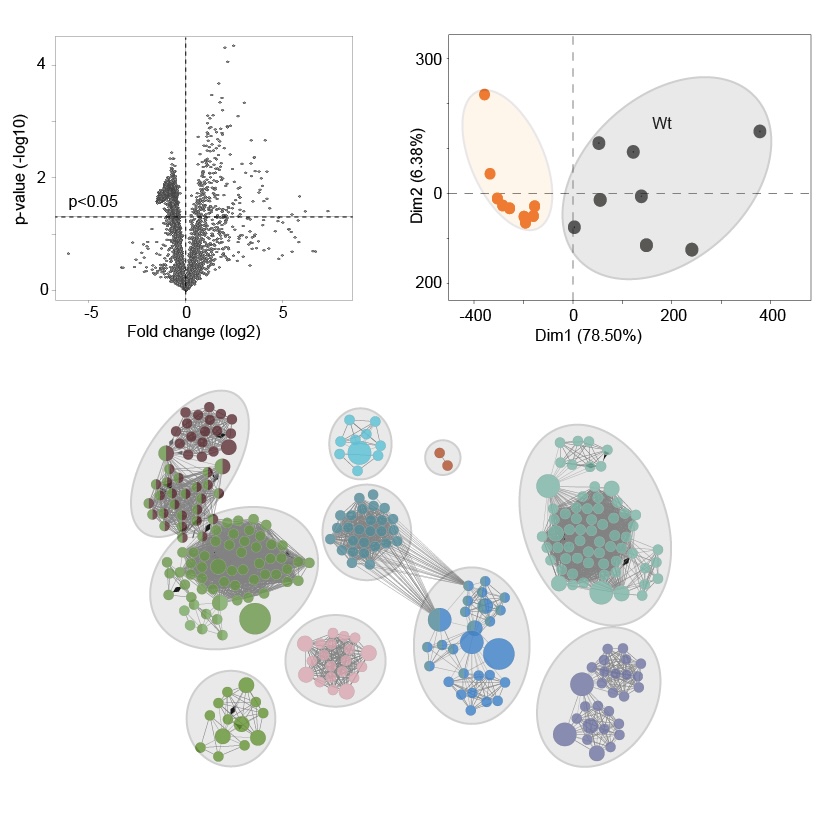
Integration of multimodal and high-dimensional datasets
The complex and high-dimensional datasets generated by mass spectrometry imaging require advanced bioinformatics approaches for meaningful interpretation. Computational methods are used to preprocess, normalize, and align spectral data, followed by multivariate statistics and machine learning to identify spatial patterns and molecular signatures. Integrative bioinformatics enables the combination of MSI data with complementary modalities such as microscopy, transcriptomics, or single-cell analyses, thereby linking molecular profiles to specific cell types and tissue structures. Through data integration and network-based interpretation, these approaches facilitate hypothesis generation, biomarker discovery, and a systems-level understanding of biological processes in health and disease.